Monitoring inland water quality in Poland using Python and Sentinel-2 satellite imagery
Substantive overview
Water quality in inland bodies of water is a critical environmental issue in Poland, primarily driven by rapid urbanization, industrial activities, and inadequate wastewater treatment infrastructure, especially in rural areas. These challenges have led to significant water pollution, resulting in detrimental effects on ecosystems, human health, and overall water resources. Traditional methods of water quality monitoring, which rely heavily on manual sampling and laboratory analysis, are not only labor-intensive but also costly and time-consuming. This is particularly problematic in rural and remote areas, where access to the necessary infrastructure is limited. In response to these challenges, our project proposes a novel, cost-effective solution for real-time monitoring of inland water quality in Poland using advanced satellite imagery and Python-based data analysis.
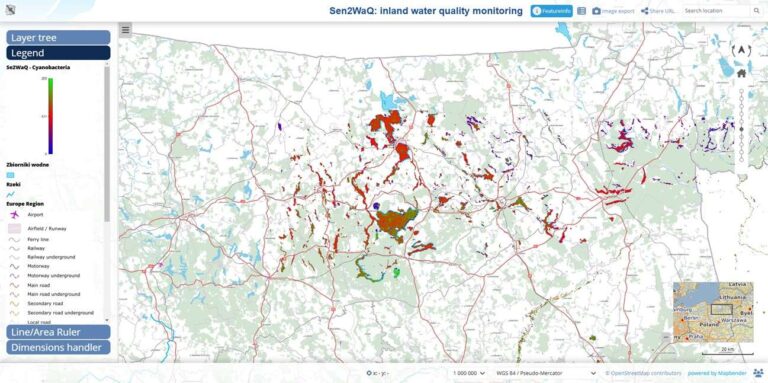
The primary goal is to develop an innovative geoportal that enables the efficient monitoring and analysis of water quality across Poland using Sentinel-2 satellite data. This approach leverages remote sensing technologies, cloud computing, and open-source tools to create a scalable system that is accessible to both researchers and the public. By utilizing satellite data, we aim to overcome the financial and logistical constraints of traditional monitoring systems, providing continuous data coverage over large geographic areas without the need for costly fieldwork.
The impetus for this project arises from the growing concerns over water pollution in Poland. Urban and industrial development has significantly increased the levels of pollutants discharged into rivers, lakes, and coastal waters. This pollution not only affects water quality but also leads to eutrophication, resulting in "oxygen deserts" where aquatic life cannot survive.
The project seeks to address these challenges by utilizing the capabilities of satellite-based remote sensing. Sentinel-2 satellites, operated by the European Space Agency, provide high-resolution multispectral imagery that is well-suited for monitoring water quality. By analyzing this data, we can derive critical water quality indicators, such as chlorophyll concentration, turbidity, and the presence of harmful algal blooms. This approach not only reduces costs but also allows for the continuous monitoring of large and remote areas, making it an ideal solution for Poland’s diverse landscape.
Technical overview
Data Acquisition
Our project is built on the integration of various technologies and platforms to create a robust system for water quality assessment. The workflow begins with the acquisition of the newest Sentinel-2 imagery through the CREODIAS platform, which provides access to a vast repository of Earth Observation data.
Data Processing
Data processing is performed on Virtual Machines provided by CREODIAS, which offer enhanced computational power for handling large datasets. Once the real-time satellite data is obtained, we process it using Python libraries such as Rasterio and GDAL.
Data Storage
The processed data, including the calculated indices, is then stored in Amazon S3 Bucket, a scalable cloud storage service that ensures efficient data management and retrieval. To maintain metadata and facilitate easy access to the results, we utilize a PostgreSQL database.
WMS and Visualization
For the visualization component, we deploy Web Map Services using MapServer. The WMS outputs are further enhanced using Mapbender, framework for creating interactive web maps. This enables users to visualize water quality data dynamically, providing an intuitive interface for exploring spatial patterns and trends over time. The final output is a user-friendly geoportal that displays water quality data in an accessible and visually engaging manner.
Summary
By making this data publicly available, we aim to prompt concern about the state of inland waters in Poland and encourage collective action to improve water management practices.
Additionally, the geoportal serves as a valuable tool for public engagement. By making water quality data easily accessible, the portal can increase awareness of environmental challenges and encourage community involvement in water conservation efforts. The visual representation of water quality data helps to translate complex scientific information into a format that is understandable by non-specialists, thereby fostering greater public concern and action.
Looking ahead, we plan to expand the capabilities of the geoportal by incorporating additional data sources, such as Landsat imagery for water temperature monitoring and in-situ measurements, to improve the accuracy and reliability of water quality assessments. We also aim to enhance the analytical capabilities of the system by integrating machine learning algorithms to gain super image resolution.
Overall, the project demonstrates the potential of remote sensing technologies in addressing pressing environmental challenges. By providing real-time and actionable data, we hope to contribute to better water management practices in Poland and beyond. Our approach serves as a model that can be adapted for other regions facing similar challenges, promoting the use of innovative technologies for environmental conservation.
This abstract, with a focus on innovation, technical robustness, and societal relevance, will contribute valuable insights into the application of remote sensing for sustainable environmental management and decision-making.





Authors: Klaudia Kościuk and Łukasz Firek from AGH STARS coordinated by PhD Michał Lupa at the AGH Space Technology Center
Contact: kkosciuk@student.agh.edu.pl, firek@student.agh.edu.pl
The project was carried out as part of the 2024 summer student internship at CloudFerro.

